- Genome sequencing is the process that involves deciphering the exact order of base pairs in an individual. This “deciphering” or reading of the genome is what sequencing is all about. Costs of sequencing differ based on the methods employed to do the reading or the accuracy stressed upon in decoding the genome.
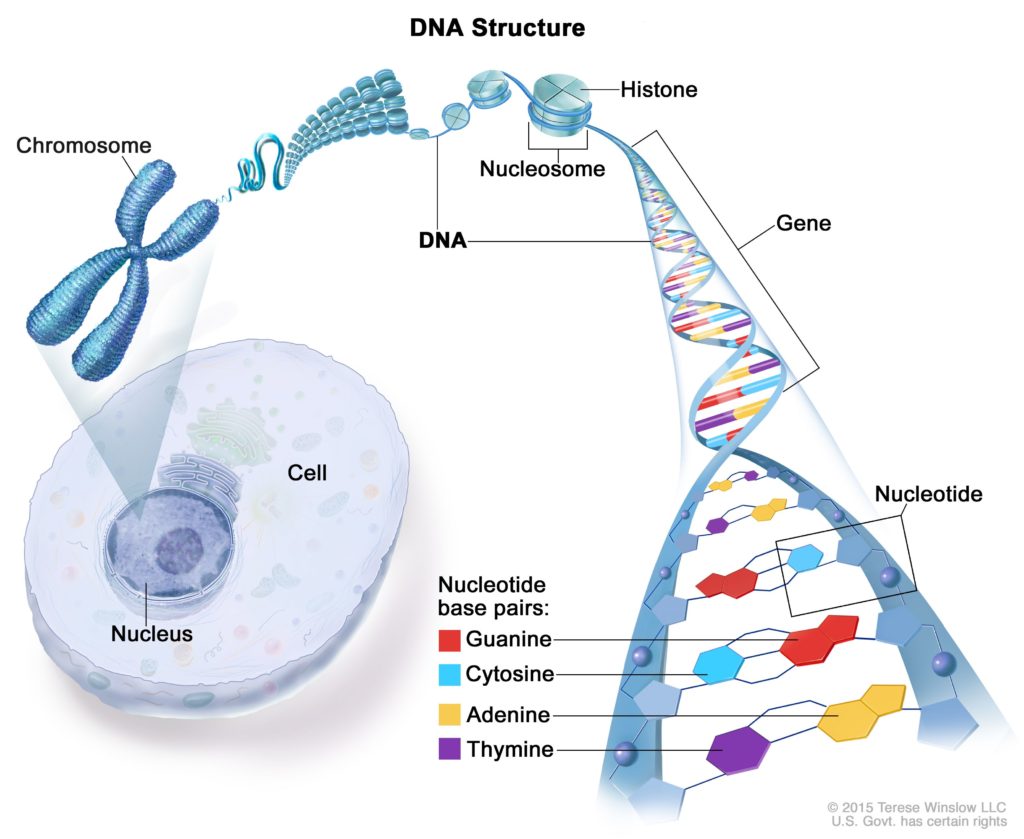
- Genome Sequencing is a laboratory process through which scientists get the complex DNA sequence (in terms of A,T,G,C) of an organism’s genome at a time.
- DNA containing cells such as saliva, epithelial cells, bone marrow, hair, seeds, and plant leaves are used as samples for sequencing.
- Genome sequencing is done by an instrument called automated DNA sequences.
Need for genome sequencing:
- Ever since the human genome was first sequenced in 2003, it opened a fresh perspective on the link between disease and the unique genetic make-up of each individual.
- Nearly 10,000 diseases — including cystic fibrosis, and thalassemia — are known to be the result of a single gene malfunctioning.
- While genes may render some insensitive to certain drugs, genome sequencing has shown that cancer too can be understood from the viewpoint of genetics, rather than being seen as a disease of certain organs
Human Genome Project (HGP)
The Human Genome Project (HGP) was an international scientific research project with the goal of determining the base pairs that make up human DNA, and of identifying and mapping all of the genes of the human genome from both a physical and a functional standpoint.
HGP and its benefits:
- Identification of predictive and predisposition markers would be accelerated by harnessing the huge amount of genetic variability available for common and complex diseases
- Data from the project is expected to answer several questions involving similarities and differences between humans and our closest relatives.
Specific Applications – Genome research could be potentially helpful in other fields
- Medicine
- Improved diagnosis of disease and earlier detection of genetic predispositions to disease
- Rational drug design
- Gene therapy
- Reduce the likelihood of heritable mutations
- Access health damage and risks caused by radiation exposure to mutagenic chemicals and cancer-causing toxins.
- Evolutionary Biology and Anthropology
- Study of evolution
- Study of migration of different human population groups
- Energy and Environment applications
- Detect bacteria and other organisms that may pollute air, water, soil and food.
- DNA Forensics
- Identify crimes and catastrophe victims.
- Match organ donors with recipients in transplant programs
- Agriculture
- Development if productive and disease, insect and drought-resistant crops
- Healthier, more productive, and disease-resistant farm animals.
National Genomic Grid
- It will study genomic data of cancer patients from India.
- It will collect samples from cancer patients, through a network of pan-India collection centres by bringing all cancer treatment institutions on board.
- The grid to be formed will be in line with the National Cancer Tissue Biobank (NCTB) set up at the Indian Indian Institute of Technology, Madras.
- National Cancer Tissue Biobank (NCTB), is a joint initiative of the Department of Science and Technology (DST), Government of India and Indian Institute of Technology, Madras.
- The biobank collects cancer tissue samples with consent from patients diagnosed with cancer.
- The aim is to provide researchers with high quality of cancer tissues and the patient data in order to facilitate cancer research that will lead to improvements in cancer diagnosis and treatment.
- This research is carried out through the technique of Genome Sequencing.
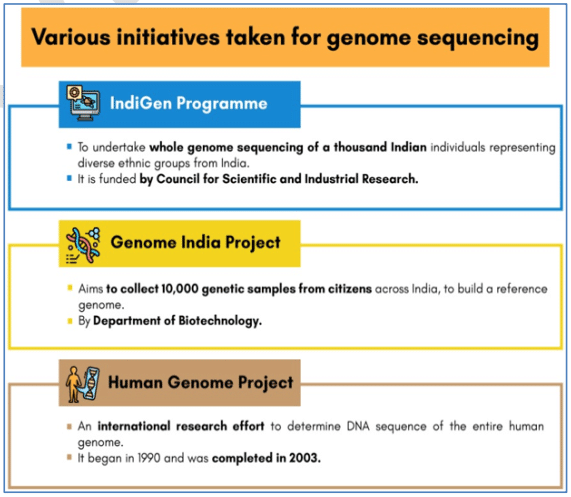
- Moreover, under Genome India Initiative, the government plans to scan 20,000 Indian genomes (in the next five years) in order to develop diagnostic tests and effective therapies for treating diseases such as cancer.
Human Genome Project-Write
- A complete haploid copy of the human genome consists of at least three billion DNA nucleotide base pairs, which have been described in the Human Genome Project – Read program (95% completed as of 2004).
- Among the many goals of GP-Write are the making of cell lines resistant to all viruses and synthesis assembly lines to test variants of unknown significance that arise in research and diagnostic sequencing of human genomes (which has been exponentially improving in cost, quality, and interpretation).
Indigen Project
- The IndiGen initiative was undertaken by CSIR in April 2019, which was implemented by the CSIR-Institute of Genomics and Integrative Biology (IGIB), Delhi and CSIR-Centre for Cellular and Molecular Biology (CCMB), Hyderabad.
- The objective is to enable genetic epidemiology and develop public health technologies applications using population genome data.
- This has enabled benchmarking the scalability of genome sequencing and computational analysis at population scale in a defined timeline.
- The ability to decode the genetic blueprint of humans through whole genome sequencing will be a major driver for biomedical science.
- IndiGen programme aims to undertake whole genome sequencing of thousands of individuals representing diverse ethnic groups from India.
Nanopore Gene Sequencing
- It enables direct, real-time analysis of long DNA or RNA fragments at a faster and cheaper rate than previously possible with older technologies.
- It works by monitoring changes to an electrical current as nucleic acids are passed through a protein nanopore.
- Nucleic acids are essential for all forms of life and are found in all cells and viruses. Nucleic acids come in two natural forms called DNA and RNA.
- The resulting signal is decoded to provide specific DNA or RNA sequence.
DNA Microarray
- In recent years, a new technology, called DNA Microarray, has attracted tremendous interest among biologists.
- This technology promises to monitor the whole genome on a single chip so that researchers can have a better picture of the interactions among thousands of genes simultaneously.
- It is widely believed that thousands of genes and their products (i.e., RNA and proteins) in a given organism function in a complicated and orchestrated way that creates the mystery of life.
- However, traditional methods in molecular biology generally work on a “one gene-one experiment” basis, which means that the throughput is very limited and the “whole picture” of gene function is hard to obtain. Microarray Technology promises to study multiple genes in one experiment.
- Microarrays consist of large numbers of DNA molecules spotted in a systematic order on a solid substrate, usually a slide. The base pairing or hybridization is the underlying principle of DNA microarray.
- Microarray exploits the preferential binding of complementary single-stranded nucleic acids.
- A microarray is typically a glass (or some other material) slide, onto which DNA molecules are attached at fixed locations (spots).
- There are several names to this technology – DNA arrays, gene chips, biochips, DNA chips, and gene arrays. The DNA microarray technology is used for analyzing the expression of thousands of messenger RNA molecules.
This technique has been used to study the following:
- Tissue-specific genes
- Regulatory gene defects in a disease
- Cellular responses to the environment
- Cell cycle variations
Gel Electrophoresis
- Gel Electrophoresis is a technique used to separate DNA fragments according to their size.
- Since DNA fragments are negatively-charged molecules, they can be separated by forcing them to move towards the anode under an electric field through a medium/matrix.
- Nowadays the most commonly used matrix is agarose which is a natural polymer extracted from seaweeds.
- The DNA fragments separate (resolve) according to their size through the sieving effect provided by the agarose gel. Hence, the smaller the fragment size, the farther it moves.
- The separated DNA fragments can be visualized only after staining the DNA with a compound known as ethidium bromide followed by exposure to UV radiation (you cannot see pure DNA fragments in the visible light and without staining)
- The separated bands of DNA are cut out from the agarose gel and extracted from the gel piece. This step is known as elution.
- The DNA fragments purified in this way are used in constructing recombinant DNA by joining them with cloning vectors.
Genetic Marker
- A short sequence of DNA with a known location on chromosome useful to identify cells, individuals or species
- To study the relationship between an inherited disease and its genetic cause, genetic markers can be used.
Biomarker
In medicine, a biomarker is a measurable indicator of the severity or presence of some disease state. More generally a biomarker is anything that can be used as an indicator of a particular disease state or some other physiological state of an organism.
A biomarker can be a substance that is introduced into an organism as a means to examine organ function or other aspects of health. For example, rubidium chloride is used in isotopic labeling to evaluate perfusion of the heart muscle. It can also be a substance whose detection indicates a particular disease state, for example, the presence of an antibody may indicate an infection.
